Home
Research
Our group investigates the biology of the bacterial cell envelope. In Gram-negative bacteria this structure comprises two membranes (the cytoplasmic and outer membranes) and the periplasmic compartment between them which contains the cell wall. We are particularly interested in how proteins and DNA are transported across and around the cell envelope.
We address these problems using a very wide range of techniques (some collaboratively) including protein biochemistry, molecular genetics, bacterial cell biology, single molecule fluorescence imaging, structural biology, and molecular simulations.
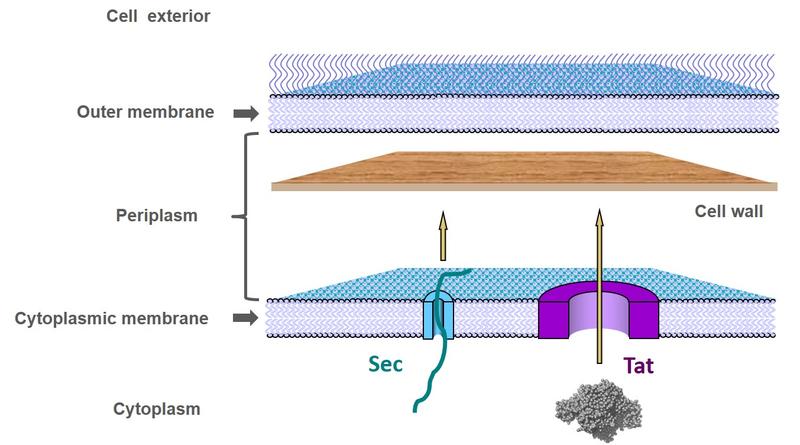
Schematic of the bacterial cell envelope showing the pathways for protein export across the cytoplasmic membrane.
Bacteria use two protein transport systems for protein export across the cytoplasmic membrane. Functionally these two systems are distinguished by the folding state of the transported protein: whilst the Sec apparatus threads unstructured proteins across the membrane, the Tat (twin-arginine translocation) system transports folded proteins.
The Tat system transports a wide range of proteins including many with bound metals or other cofactors that are required for respiratory or photosynthetic energy generation. It is essential for the virulence of bacterial pathogens. Tat is evolutionarily conserved in plant chloroplasts where it is needed for the assembly of functional photosynthetic complexes.
A particular mechanistic challenge for the Tat system is that the diverse shapes and sizes of folded proteins make it difficult to maintain the membrane seal around the transporting protein. Our laboratory seeks to understand the molecular basis of this unusual transport system.
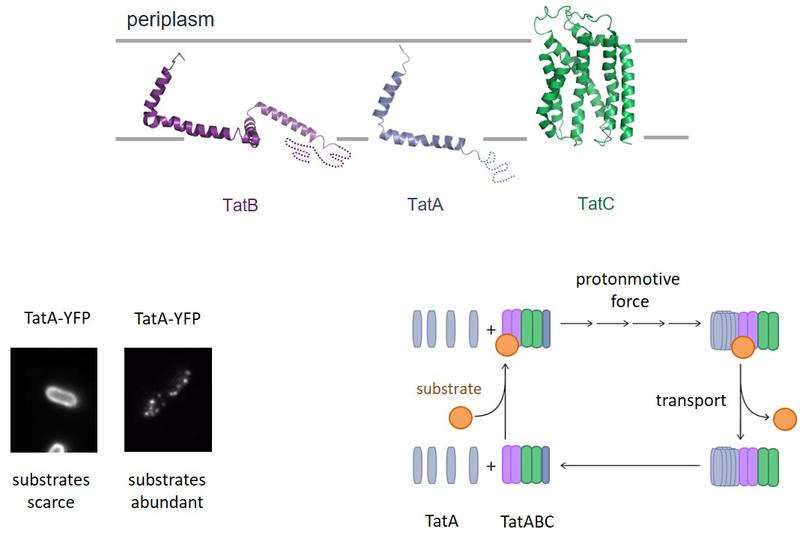
The Tat protein transport system. (Top) Structures of the individual Tat system proteins and their orientation in the bacterial cytoplasmic membrane. (Bottom Left) Live cell fluorescence imaging of a TatA-YFP (yellow fluorescent protein) fusion showing that in the presence of transport substrates TatA assembles into large oligomers visible as distinct fluorescent spots. (Bottom Right) Model for the Tat protein transport cycle. Substrate proteins bind to a TatABC complex. This triggers recruitment of multiple additional TatA molecules to form the active transport site. This process requires the transmembrane protonmotive force. Following transport of the substrate protein across the membrane the TatA oligomer disassembles. The number of Tat subunits depicted in each complex is arbitrary and was selected for illustrative convenience.
- Palmer T, and Berks BC (2012) The twin-arginine translocation (Tat) protein export pathway. Nat Rev Microbiol 10: 483-496. https://www.nature.com/articles/nrmicro2814
- Berks BC (2015) The twin-arginine protein translocation pathway. Annu Rev Biochem 84: 843-864. https://www.annualreviews.org/doi/full/10.1146/annurev-biochem-060614-034251
- Rollauer, S.E., Tarry, M.J., Graham, J.E., Jääskeläinen, M., Jäger, F., Johnson, S., Krehenbrink, M., Liu, S.-M., Lukey, M.J., Marcoux, J., McDowell, M.A., Roversi, P., Stansfeld, P.J., Robinson, C.V., Sansom, M.S.P., Palmer, T., Högbom, M., Berks, B.C., and Lea, S.M. (2012) Structure of the TatC core of the twin arginine protein transport system. Nature 492: 210-214. https://www.nature.com/articles/nature11683
- Rodriguez, F., Rouse, S.L., Tait, C., Harmer, J., de Riso, A., Timmel, C.R., Sansom, M.S.P., Berks, B.C., and Schnell, J.R. (2013) Structural model for the protein-translocating element of the twin-arginine transport system. Proc Natl Acad Sci. USA 110: E1092–E1101. https://www.pnas.org/content/110/12/E1092.long
- Alcock, F., Baker, M.A.B., Greene, N.P., Palmer, T., Wallace, M.I., and Berks, B.C. (2013) Live cell imaging shows reversible assembly of the TatA component of the twin-arginine protein transport system. Proc Natl Acad Sci USA 110: E3650–E3659. https://www.pnas.org/content/110/38/E3650.long
- Alcock, F., Stansfeld, P.J., Basit, H., Habersetzer, J., Baker, M.A.B., Palmer, T., Wallace, M.I., and Berks, B.C. (2016) Assembling the Tat protein translocase. eLife 5: e20718. https://elifesciences.org/articles/20718
Protein transport: Type 9 Secretion System
The recently discovered Type 9 Secretion System (T9SS) exports proteins across the outer membrane of bacteria of the Bacteroidetes superphylum. The T9SS is an essential virulence determinant in the human oral pathogens Porphyromonas gingivalis and Tannerella forsythia which are the causative agents of severe periodontal disease. It is also essential for the unusual rapid gliding motility found in Bacteroidetes bacteria.
The T9SS contains at least 20 protein components, the functions of which include forming the translocon (the protein conducting channel), post-transport modification of the substrate protein, and energization of transport using the protonmotive force across the cytoplasmic membrane. We study all parts of the pathway with the aim of elucidating the molecular mechanism of Type 9 transport.
PorV complex
Plug complex
Structures of two different Type 9 translocon complexes. The translocon channel protein is shown in rainbow colours and the partner proteins in grey.
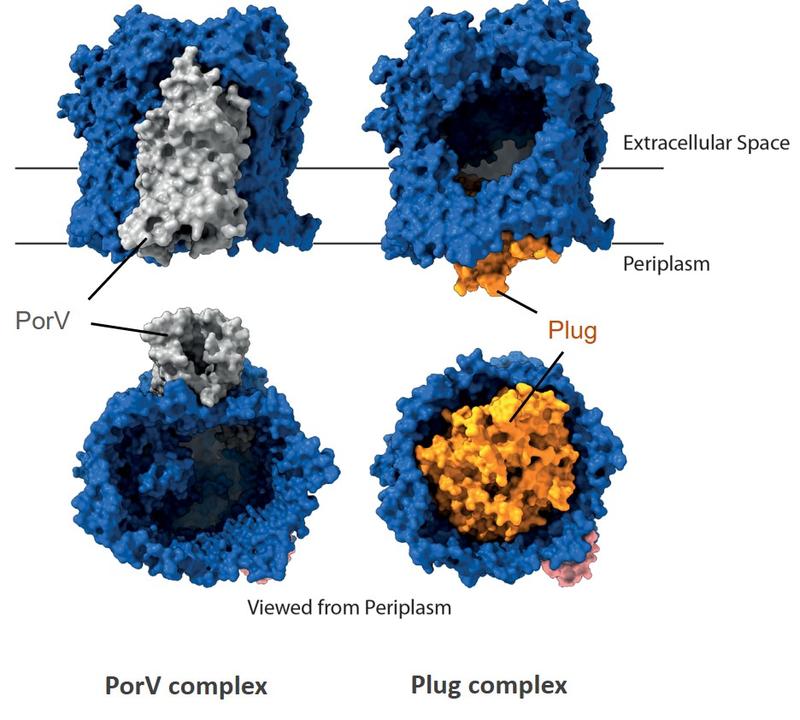
The T9SS translocon channel is alternately gated by partner proteins. The translocon channel (blue) is capped on the extracellular end but has a lateral opening to the external membrane surface. Structures of the channel protein bound to different T9SS components (PorV and Plug) demonstrate that partner proteins control access to the lateral opening and to the periplasmic end of the channel. This suggests an alternating access mechanism in which the two ends of the protein conducting channel are open at different times.
The T9SS is energised by a trans-envelope rotary motor. Proton flow through the cytoplasmic membrane segment of the complex (coloured) drives rotation of a periplasm-spanning arm (grey) to power transport at the outer membrane.
- Lauber, F., Deme, J.C, Lea, S.M., and Berks, B.C. (2018) Type 9 secretion system structures reveal a new protein transport mechanism. Nature 564: 77-82. https://www.nature.com/articles/s41586-018-0693-y
- Hennell James R, Deme JC, Kjӕr A, Alcock F, Silale A, Lauber F, Johnson S, Berks BC, and Lea SM (2021) Structure and mechanism of the proton-driven motor that powers type 9 secretion and gliding motility. Nat Microbiol 6: 221-233. https://www.nature.com/articles/s41564-020-00823-6
Gliding motility
Bacteria in the Bacteroidetes superphylum are able to move at high speed across solid surfaces using a phylum-specific gliding motility apparatus. This gliding motility allows the bacteria to travel their own cell length in a second which is a hundred times faster than eukaryotic cells move over surfaces. Gliding is mediated by the helical movement of adhesin molecules along the cell surface. We are interested in the molecular basis of Bacteroidetes gliding motility and how motility is controlled in response to external stimuli.
Bacteria gliding on glass imaged in real time.
The corkscrewing motion of gliding bacterial cells. The cell motion is revealed by imaging a fluorescently labelled protein that has a fixed position on the cell surface. The movie plays in real time.
A single fluorescently labelled adhesin molecule moving along the surface of an immobilised cell. The pathway of the adhesin is plotted in red. The movie plays in real time.
- Hennell James R, Deme JC, Kjӕr A, Alcock F, Silale A, Lauber F, Johnson S, Berks BC, and Lea SM (2021) Structure and mechanism of the proton-driven motor that powers type 9 secretion and gliding motility. Nat Microbiol 6: 221-233. https://www.nature.com/articles/s41564-020-00823-6
DNA transport in horizontal gene transfer
Horizontal gene transfer is the major route by which antibiotic resistance genes and other adaptive traits move between bacteria. Horizontal gene transfer requires the transport of DNA molecules across the cell envelope. We study the molecular mechanisms by which this occurs.
- In transformation (also termed natural competence) the bacterium takes up DNA directly from the environment.
- In conjugation a plasmid or other genetic element in a donor bacterium is transferred to a recipient bacterium through a mating bridge between the two cells.
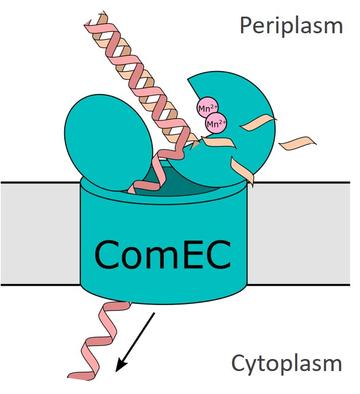
Cartoon showing the possible mechanism of DNA transport across the cytoplasmic membrane during natural transformation. The transport protein ComEC digests one strand of the double stranded DNA molecule at the periplasmic side of the membrane and transports the remaining strand through a pore to the cytoplasm.
- Silale A, Lea SM, Berks BC (2021) The DNA transporter ComEC has metal-dependent nuclease activity that is important for natural transformation. Mol Microbiol 116: 416-426. https://onlinelibrary.wiley.com/doi/10.1111/mmi.14720
Some of our collaborators
- Susan Lea (structural biology) https://ccr.cancer.gov/staff-directory/susan-m-lea
- Tracy Palmer (Tat system) https://www.ncl.ac.uk/medical-sciences/people/profile/tracypalmer.html
- Phillip Stansfeld (molecular simulations) https://warwick.ac.uk/fac/sci/lifesci/people/pstansfeld/
- Achillefs Kapanidis (single molecule fluorescence) https://kapanidis.web.ox.ac.uk/home
- Richard Berry (bacterial motility) https://www.physics.ox.ac.uk/our-people/berryr
- Kevin Foster (bacterial behaviour) https://zoo-kfoster.zoo.ox.ac.uk
- William (Mac) Durham (bacterial behaviour) https://www.sheffield.ac.uk/physics/people/academic/william-mack-durham


